Using RStudio via OnDemand¶
RStudio is a code development environment for R. It provides an intuitive web interface for developing your R workflow interactively. Using RStudio on Apocrita via OnDemand allows you to scale to much larger datasets and workloads without putting a strain on your local machine. Please refer to the overview section for instructions on how to login to OnDemand.
Starting an RStudio session¶
Select RStudio Server from the Servers list of the Interactive Apps drop-down menu, or from the My Interactive Sessions page.
Choose the resources your job will need. Choosing a 1 hour maximum running time is the best option for getting a session quickly, unless you have access to owned nodes which may offer sessions immediately for up to 24hrs if resources are available.
Choose your core request wisely
R will only use multiple cores if your code calls an appropriate
parallelisation library such as parallel or parallelly. Please ensure
your code is parallelised in short test sessions before requesting a large
number of cores for a long runtime.
Long-running sessions requesting a large core count without using these effectively may be terminated by the ITSR admin team.
Choose your RAM request wisely
Please establish a baseline of expected RAM usage in short test sessions before requesting a large amount of RAM for a long runtime.
Long-running sessions requesting a large amount of RAM without using this effectively may be terminated by the ITSR admin team.
Once clicking Launch, the request will be queued, and when resources have been allocated, you will be presented with the option to connect to the session by clicking on the blue Connect to RStudio Server button.
Once connected, the familiar RStudio interface is presented, and you will be able to use the allocated resources, and access your research data located on Apocrita.
Installing packages¶
Think twice before using renv!
The renv project manager has
become popular, but we strongly advise users to think twice before using it
on Apocrita. It aggressively caches existing binaries, but sometimes the
cached binaries for personal installs aren't compatible with all
environments and can cause issues. We have tested renv fairly extensively
a number of times, and there are still quite a few issues that need to be
resolved before we would recommend (or support) its use.
If you still choose to use renv on Apocrita despite this, the ITSR team
can't really offer any support in doing so. There are details for using
alternative locations for your libraries on Apocrita
in our main
R documentation.
It's likely your scripts will require additional R libraries; these can be
installed using the install.packages() command in the console window of
RStudio. If the package cannot be installed due to missing dependencies, please
contact us and we will try to add
them to the
Spack environment that is activated
alongside RStudio.
Further tips and guidance regarding R package installations can be found on our blog: Untangle installations
Known issues¶
Web browser security¶
For security reasons, modern browsers will restrict OnDemand from embedding certain content in the RStudio application.
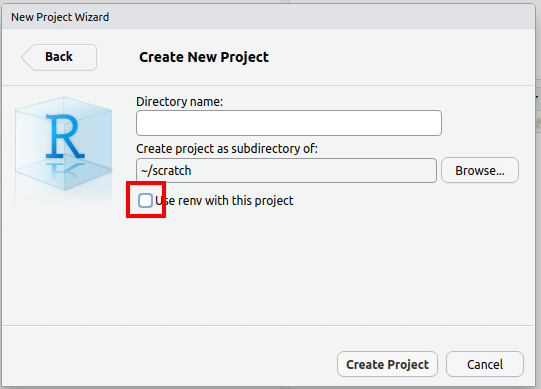
Rprojects and renv¶
As detailed above, the use of renv is not recommended.
If you use the Rproject feature of RStudio, the default is to use renv as
well. We recommend un-checking this box:
Exiting the session¶
If a session exceeds the requested running time, it will be killed. You may receive a message "The previous R session was abnormally terminated...". Click OK to acknowledge the message and continue. To avoid this message, it's good practice to exit the session cleanly when you have finished.
To cleanly exit RStudio, click File -> Quit Session... and then release resources back to the cluster queues by clicking the red Delete button for the relevant session on the My Interactive Sessions page.
For further guidance on exiting RStudio sessions please see this section of our blog: Exit intentionally
Useful additional tips¶
Further tips regarding the usage of RStudio can be found under our blog site:
R Top Tips